Contact Information, Courses, Grants, Publications, etc.
 Studies on latent DNA replication and long-term episomal maintenance of KSHV/HHV-8
Studies on latent DNA replication and long-term episomal maintenance of KSHV/HHV-8
 Kaposi’s sarcoma-associated herpesvirus (KSHV) also called Human herpes virus type 8 (HHV-8) is associated with Kaposis’s sarcoma (KS), and two lymphoproliferative diseases: Primary effusion lymphomas (PEL) and a subset of Multicentric Castlemen’s disease (MCD). Common to these malignancies is that the majority of tumor cells are latently infected and express only a small number of viral genes.
Kaposi’s sarcoma-associated herpesvirus (KSHV) also called Human herpes virus type 8 (HHV-8) is associated with Kaposis’s sarcoma (KS), and two lymphoproliferative diseases: Primary effusion lymphomas (PEL) and a subset of Multicentric Castlemen’s disease (MCD). Common to these malignancies is that the majority of tumor cells are latently infected and express only a small number of viral genes.
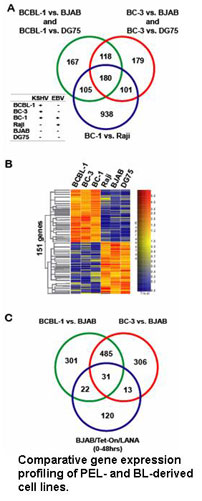
One of these genes, ORF73, encodes the latency-associated nuclear antigen (LANA). LANA is highly expressed in all KS tumor cells and plays an important role in the biology of KSHV. LANA is the only viral protein required for latent DNA replication and genome maintenance during latency. In addition, LANA interacts with a variety of cellular proteins including the tumor suppressors p53 and Rb thereby 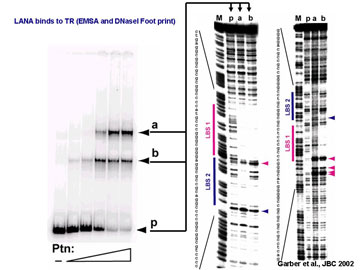 modulating gene expression in latently infected cells. To analyze LANA’s ability to modulate cellular gene expression, we have performed gene expression profiling and found that LANA regulates a number of genes in the Rb/E2F pathway which, as a consequence can protect lymphoid cells from p161NK4a-induced cell cycle arrest.
modulating gene expression in latently infected cells. To analyze LANA’s ability to modulate cellular gene expression, we have performed gene expression profiling and found that LANA regulates a number of genes in the Rb/E2F pathway which, as a consequence can protect lymphoid cells from p161NK4a-induced cell cycle arrest.
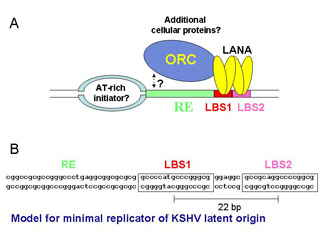
To study LANA’s role in DNA replication we performed a detailed biochemical analysis of its DNA binding specificity and activity. We identified two binding sites within the terminal repeat of the KSHV genome and demonstrated that both sites contribute to the ability of LANA to support replication of TR-containing plasmids.
 We hypothesize that LANA and/or cis-regulatory sequences within TR interact with cellular proteins that facilitate DNA replication. We are currently analyzing the mechanisms by which LANA supports DNA replication by: 1) Mapping cis-regulatory sequences of the putative origin of replication within TR 2) Mapping trans-requirements for DNA binding and LANA-dependent DNA replication 3) Identification of cellular proteins that interact with LANA-C and/or a minimal origin of replication 4) Understanding how the cellular environment contributes to the establishment and maintenance of viral latency.
We hypothesize that LANA and/or cis-regulatory sequences within TR interact with cellular proteins that facilitate DNA replication. We are currently analyzing the mechanisms by which LANA supports DNA replication by: 1) Mapping cis-regulatory sequences of the putative origin of replication within TR 2) Mapping trans-requirements for DNA binding and LANA-dependent DNA replication 3) Identification of cellular proteins that interact with LANA-C and/or a minimal origin of replication 4) Understanding how the cellular environment contributes to the establishment and maintenance of viral latency.
Based on the fact that LANA is required not only for long-term maintenance but also for DNA replication, we hypothesize that LANA is an attractive target for the development of antiviral compounds. Such potential drugs would specifically act on latently infected cells and conceivably, could also prevent the establishment of latency in de novo infected cells. To directly test our hypothesis, we have developed a fluorescence anisotropy-based high-throughput screening assay (HTS) to identify small molecule inhibitors of LANA function. To validate potential candidates we are developing tissue culture-based assays which monitor long-term maintenance.
The long-term goal of our studies is to gain is to increase our knowledge on the mechanisms by which KSHV successfully establish and maintain latency – potentially these studies can lead to the development of novel therapeutic strategies.
Functional analysis of virally-encoded microRNAs. Most recently, our laboratory has started working on KSHV-encoded microRNAs (miRNAs). MiRNAs represent a novel class of post-transcriptional regulators, which bind to 3, UTRs of mRNAs and either 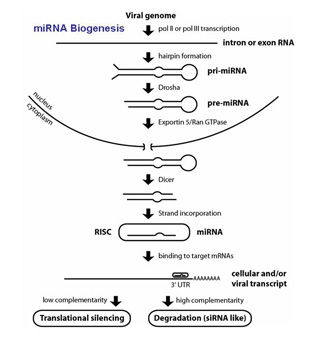 slow down their translation and/or induce their degradation. It is now known that a large number of DNA viruses also encode miRNAs their by likely modulate the host cellular transcriptome. We have cloned 11 KSHV miRNAs and now are pursuing research that address the question how and if these RNA regulators contribute to the biology of KSHV. Indications that these novel viral regulators may play important roles in viral biology come from the fact that they are coordinately expressed with the latency-associated genes and that they are highly conserved among virus isolates. We have performed RNA mapping studies and analyzed more than 30 clinical isolates from all over the world. This data strongly suggests that miRNA genes are selected for in vivo.
slow down their translation and/or induce their degradation. It is now known that a large number of DNA viruses also encode miRNAs their by likely modulate the host cellular transcriptome. We have cloned 11 KSHV miRNAs and now are pursuing research that address the question how and if these RNA regulators contribute to the biology of KSHV. Indications that these novel viral regulators may play important roles in viral biology come from the fact that they are coordinately expressed with the latency-associated genes and that they are highly conserved among virus isolates. We have performed RNA mapping studies and analyzed more than 30 clinical isolates from all over the world. This data strongly suggests that miRNA genes are selected for in vivo.
Currently, we are pursuing a variety of genomics, proteomics, and ribonomics-based approaches to determine host cellular and/or viral genes that may be regulated by KSHV-encoded miRNAs.
Samols, S., Hu, J., Skalsky, R., and R. Renne. Cloning and identification of a microRNA cluster in the latency-associated region of Kaposi’s sarcoma-associated herpesvirus. Journal of Virology. 2005. 79(14): 9301-05.
Samols, M and R. Renne. Virus Encoded MicroRNAs: A new chapter of virus/host cell interactions. Future Virology 2006 1(2) 233-242.
An, F., Compitello, N., Folarin, H., Roth, J., Gerson, SL., Mc Crae, K., and R. Renne. Telomerase-immortalized endothelial cells: a model system to study KSHV infection, latency, and tumorigenesis in vivo. Journal of Virology 2006. 80(10):4833-46. “JVI Spotlight”
Hu, J., Samols, M.S., Marshall, V., Parks, T., Wang, C.D., Whitby, D., and R. Renne Expression and conservation of Kaposi’s sarcoma-associated herpesvirus encoded microRNAs. Submitted for publication.
Skalsky, R.L., Hu, J., and R. Renne. Modelling episomal maintenance of Kaposi’s sarcoma-associated herpesvirus utilizing GFP reporter replicons. Submitted for publication.
Education:
Postdoctoral fellow, University of California, San Francisco
Ph.D. Albert-Ludwigs University, Freiburg, Germany 1993
Diploma. Albert-Ludwigs University, Freiburg,Germany 1989
Awards and Professional Services:
Leukemia Society of America, Special Fellow 1997-2000
Mt. Sinai Health Care Foundation, Cleveland, Scholar Award 1998-2000
Member NIH AOIC Study section
![]() Citations
Citations
Teaching Responsibilities:
GMS 7979 Advanced Research
GMS 7980 Doctoral Research

